Plant Disease Detection Using Image Segmentation
Sethi M.1*, Singh P.2
DOI: 10.54060/ijahr.v1i1.3
1* Mohit Sethi, Student, Department of Computer Science & Engineering, Amity School of Engineering & Technology Lucknow, Amity University Uttar Pradesh, Lucknow, Uttar Pradesh, India.
2 Pawan Singh, Associate Professor, Department of Computer Science & Engineering, Amity School of Engineering & Technology Lucknow, Amity University Uttar Pradesh, Lucknow, Uttar Pradesh, India.
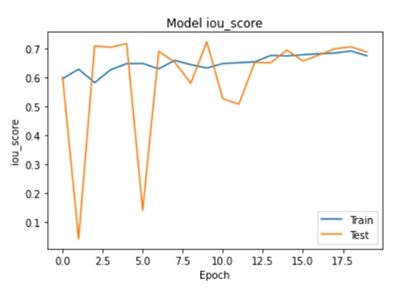
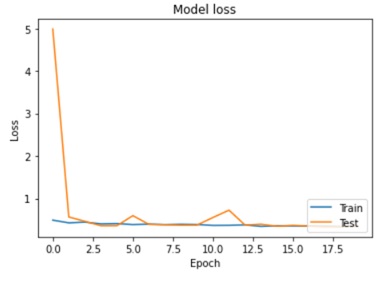
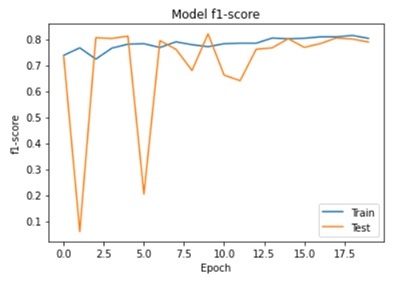
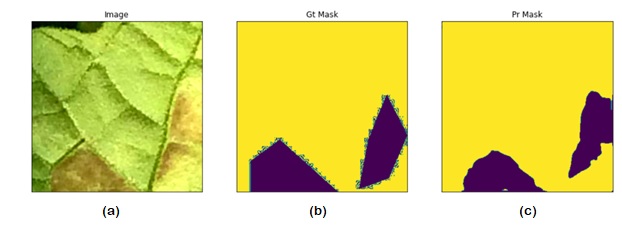
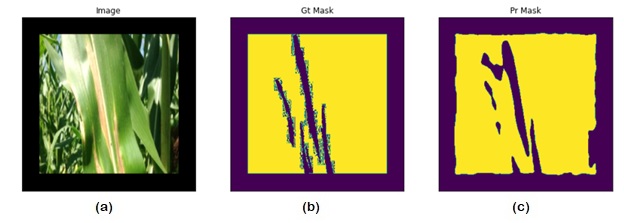
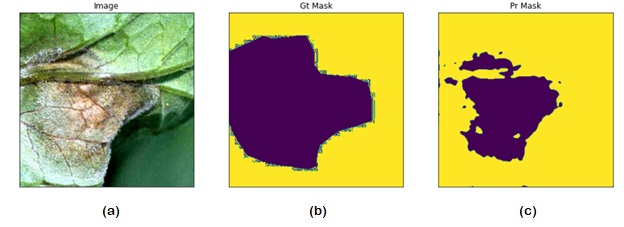
Abstract: This paper presents a novel approach for detecting plant diseases using image segmentation techniques. The proposed method employs deep learning algorithms to segment images into healthy and infected areas, and then classifies the disease based on the segmented region. The use of image segmentation allows for the automated detection and quantification of diseases in plants, making it a valuable tool for farmers and researchers. Experimental results show that the proposed method achieves high accuracy in detecting various plant diseases, including leaf spot, powdery mildew, and rust. The method's performance was evaluated on a dataset of plant images, demonstrating its effectiveness in real-world applications. The proposed approach has the potential to revolutionize the way plant diseases are detected and managed, improving crop yields and reducing losses due to disease outbreaks.
Keywords: Deep Learning, Image Segmentation, Sematic Segmentation, Transfer Learning, Case Report
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , Student, Department of Computer Science & Engineering, Amity School of Engineering & Technology Lucknow, Amity University Uttar Pradesh, Lucknow, Uttar Pradesh, India. Email: |
Mohit Sethi, Pawan Singh, Plant Disease Detection Using Image Segmentation. IJAHR. 2023;1(1):15-18. Available From https://ahr.a2zjournals.com/index.php/ahr/article/view/3/version/3 |


 ©
©